Read this blog in:
Antibiotic resistance imparted by the versatile Pseudomonas aeruginosa
Pseudomonas aeruginosa is a Gram negative bacterial species which has been identified as an opportunistic pathogen responsible for causing infections in blood, lungs or other parts of the body. In 2017, multidrug-resistant (MDR) P. aeruginosa caused an estimated 32,600 infections among hospitalized patients and 2,700 estimated deaths in the United States only [Source: 2019 AR Threats Report].
This MDR “superbug” P. aeruginosa can still be treated with classical antibiotics. But owing to the rapid increase in the scope of the resistance demonstrated by the bacteria to various antibiotics, it has become difficult to choose the right antibiotic for treatment during an infection. As a result, doctors usually advice to send a specimen for culture to the laboratory and for testing against various antibiotics.
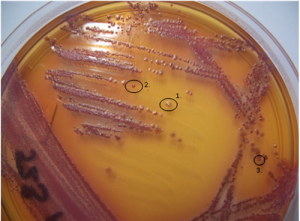
Morphological variation of P. aeruginosa isolates from culture of a CF sputum sample.(Source: L Laine et al 2009)
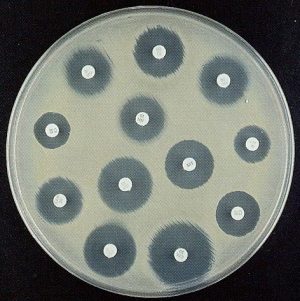
Assays used for AST: 1) Disc diffusion (Source: Google images)

Assays used for AST: 2) ETEST (Source: Google images)
Susceptibility testing
The routine diagnostic procedure for antibiotic susceptibility testing (AST) involves procurement of a relevant clinical sample; letting the bacteria grow on conventional culture media for approximately 24 hours; using specific laboratory assays to determine the MIC (minimum inhibitory concentration). For completion of these steps an average of 48 hours is needed before the medical practitioner can actually be advised which antibiotic would suit best for the treatment. This should also include a targeted advice for the dosage to be applied. Thanks to the advancement in the technology, automated susceptibility testing can be done and the results could possibly be obtained within a day. Through my ongoing project, I am trying to determine which assays / technologies could be used to ascertain the correct AST profiling for MDR P. aeruginosa and could also reduce the time to result without compromising with the accuracy of the results.
For my study I received P. aeruginosa strains from the Freeman hospital in Newcastle upon Tyne, UK. Unlike the P. aeruginosa strains I had dealt with earlier, these strains were surprisingly slow growing and mucoid. As a result, the usual protocol to determine AST had to be changed. I tested the strains against a range of antibiotics using different AST assays only to find out that almost all the strains were multi-drug resistant (MDR).
Collaborations
In order to exchange data and know the progress of various projects, a meeting between bioMérieux and the Freeman hospital is conducted twice every year. Last year I attended the meeting in Newcastle upon Tyne in May. I introduced the collaborators to ViBrANT and to my project. It was indeed a nice experience for me as I got their advice on how to cultivate the slow growing strains. They also shared with me the assays they use regularly in hospital settings to determine MICs. I realized how important it is to determine those MICs especially when administering antibiotics to the patients from intensive care units or to those who are immunocompromised.
The over or under use of antibiotics is what may possibly render the pathogens more resistant. They adapt themselves very quickly rendering the antibiotics useless. According to the recent report of the CDC, 33.9% of P. aeruginosa isolates were resistant to at least one of the antimicrobial groups under surveillance (piperacillin ± tazobactam, fluoroquinolones, ceftazidime, aminoglycosides and carbapenems). This is alarming!
I am currently also studying the antibiotic resistance genes in the P. aeruginosa genomes that are responsible for imparting such high resistance to a wide array of antibiotics. Using various publicly available databases I am trying to find the differences at the genomic level between the MDR strains and the non-MDR strains.
The more I am studying this bacterial species the more I am getting fascinated by its ability to adapt to environmental conditions and to emerge victorious in the presence of different antibiotics. The next challenge for me is to quantify the biofilms produced by the strains I am studying. I am currently having a hard time doing it. In this quest of studying the biofilms produced by the versatile and adamant P. aeruginosa, I want to emerge victorious. 🙂

Process of biofilm formation (Source : Google images)



